Archive for the ‘Free software’ Category
I’m working on optimising some R code written by a researcher at University of Sheffield and its very much a war of attrition! There’s no easily optimisable hotspot and there’s no obvious way to leverage parallelism. Progress is being made by steadily identifying places here and there where we can do a little better. 10% here and 20% there can eventually add up to something worth shouting about.
One such micro-optimisation we discovered involved multiplying two matrices together where one of them needed to be transposed. Here’s a minimal example.
#Set random seed for reproducibility set.seed(3) # Generate two random n by n matrices n = 10 a = matrix(runif(n*n,0,1),n,n) b = matrix(runif(n*n,0,1),n,n) # Multiply the matrix a by the transpose of b c = a %*% t(b)
When the speed of linear algebra computations are an issue in R, it makes sense to use a version that is linked to a fast implementation of BLAS and LAPACK and we are already doing that on our HPC system.
Here, I am using version 3.3.3 of Microsoft R Open which links to Intel’s MKL (an implementation of BLAS and LAPACK) on a Windows laptop.
In R, there is another way to do the computation c = a %*% t(b) — we can make use of the tcrossprod function (There is also a crossprod function for when you want to do t(a) %*% b)
c_new = tcrossprod(a,b)
Let’s check for equality
c_new == c [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [1,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [2,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [3,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [4,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [5,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [6,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [7,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [8,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [9,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE [10,] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
Sometimes, when comparing the two methods you may find that some of those entries are FALSE which may worry you!
If that happens, computing the difference between the two results should convince you that all is OK and that the differences are just because of numerical noise. This happens sometimes when dealing with floating point arithmetic (For example, see https://www.walkingrandomly.com/?p=5380).
Let’s time the two methods using the microbenchmark package.
install.packages('microbenchmark')
library(microbenchmark)
We time just the matrix multiplication part of the code above:
microbenchmark( original = a %*% t(b), tcrossprod = tcrossprod(a,b) ) Unit: nanoseconds expr min lq mean median uq max neval original 2918 3283 3491.312 3283 3647 18599 1000 tcrossprod 365 730 756.278 730 730 10576 1000
We are only saving microseconds here but that’s more than a factor of 4 speed-up in this small matrix case. If that computation is being performed a lot in a tight loop (and for our real application, it was), it can add up to quite a difference.
As the matrices get bigger, the speed-benefit in percentage terms gets lower but tcrossprod always seems to be the faster method. For example, here are the results for 1000 x 1000 matrices
#Set random seed for reproducibility set.seed(3) # Generate two random n by n matrices n = 1000 a = matrix(runif(n*n,0,1),n,n) b = matrix(runif(n*n,0,1),n,n) microbenchmark( original = a %*% t(b), tcrossprod = tcrossprod(a,b) ) Unit: milliseconds expr min lq mean median uq max neval original 18.93015 26.65027 31.55521 29.17599 31.90593 71.95318 100 tcrossprod 13.27372 18.76386 24.12531 21.68015 23.71739 61.65373 100
The cost of not using an optimised version of BLAS and LAPACK
While writing this blog post, I accidentally used the CRAN version of R. The recently released version 3.4. Unlike Microsoft R Open, this is not linked to the Intel MKL and so matrix multiplication is rather slower.
For our original 10 x 10 matrix example we have:
library(microbenchmark)
#Set random seed for reproducibility
set.seed(3)
# Generate two random n by n matrices
n = 10
a = matrix(runif(n*n,0,1),n,n)
b = matrix(runif(n*n,0,1),n,n)
microbenchmark(
original = a %*% t(b),
tcrossprod = tcrossprod(a,b)
)
Unit: microseconds
expr min lq mean median uq max neval
original 3.647 3.648 4.22727 4.012 4.1945 22.611 100
tcrossprod 1.094 1.459 1.52494 1.459 1.4600 3.282 100
Everything is a little slower as you might expect and the conclusion of this article — tcrossprod(a,b) is faster than a %*% t(b) — seems to still be valid.
However, when we move to 1000 x 1000 matrices, this changes
library(microbenchmark)
#Set random seed for reproducibility
set.seed(3)
# Generate two random n by n matrices
n = 1000
a = matrix(runif(n*n,0,1),n,n)
b = matrix(runif(n*n,0,1),n,n)
microbenchmark(
original = a %*% t(b),
tcrossprod = tcrossprod(a,b)
)
Unit: milliseconds
expr min lq mean median uq max neval
original 546.6008 587.1680 634.7154 602.6745 658.2387 957.5995 100
tcrossprod 560.4784 614.9787 658.3069 634.7664 685.8005 1013.2289 100
As expected, both results are much slower than when using the Intel MKL-lined version of R (~600 milliseconds vs ~31 milliseconds) — nothing new there. More disappointingly, however, is that now tcrossprod is slightly slower than explicitly taking the transpose.
As such, this particular micro-optimisation might not be as effective as we might like for all versions of R.
For a while now, Microsoft have provided a free Jupyter Notebook service on Microsoft Azure. At the moment they provide compute kernels for Python, R and F# providing up to 4Gb of memory per session. Anyone with a Microsoft account can upload their own notebooks, share notebooks with others and start computing or doing data science for free.
They University of Cambridge uses them for teaching, and they’ve also been used by the LIGO people (gravitational waves) for dissemination purposes.
This got me wondering. How much power does Microsoft provide for free within these notebooks? Computing is pretty cheap these days what with the Raspberry Pi and so on but what do you get for nothing? The memory limit is 4GB but how about the computational power?
To find out, I created a simple benchmark notebook that finds out how quickly a computer multiplies matrices together of various sizes.
- The benchmark notebook is here on Azure https://notebooks.azure.com/walkingrandomly/libraries/MatrixMatrix
- and here on GitHub https://github.com/mikecroucher/Jupyter-Matrix-Matrix
Matrix-Matrix multiplication is often used as a benchmark because it’s a common operation in many scientific domains and it has been optimised to within an inch of it’s life. I have lost count of the number of times where my contribution to a researcher’s computational workflow has amounted to little more than ‘don’t multiply matrices together like that, do it like this…it’s much faster’
So how do Azure notebooks perform when doing this important operation? It turns out that they max out at 263 Gigaflops! 
For context, here are some other results:
- A 16 core Intel Xeon E5-2630 v3 node running on Sheffield’s HPC system achieved around 500 Gigaflops.
- My mid-2014 Mabook Pro, with a Haswell Intel CPU hit, hit 169 Gigaflops.
- My Dell XPS9560 laptop, with a Kaby Lake Intel CPU, manages 153 Gigaflops.
As you can see, we are getting quite a lot of compute power for nothing from Azure Notebooks. Of course, one of the limiting factors of the free notebook service is that we are limited to 4GB of RAM but that was more than I had on my own laptops until 2011 and I got along just fine.
Another fun fact is that according to https://www.top500.org/statistics/perfdevel/, 263 Gigaflops would have made it the fastest computer in the world until 1994. It would have stayed in the top 500 supercomputers of the world until June 2003 [1].
Not bad for free!
[1] The top 500 list is compiled using a different benchmark called LINPACK so a direct comparison isn’t strictly valid…I’m using a little poetic license here.
There are lots of Widgets in ipywidgets. Here’s how to list them
from ipywidgets import * widget.Widget.widget_types
At the time of writing, this gave me
{'Jupyter.Accordion': ipywidgets.widgets.widget_selectioncontainer.Accordion,
'Jupyter.BoundedFloatText': ipywidgets.widgets.widget_float.BoundedFloatText,
'Jupyter.BoundedIntText': ipywidgets.widgets.widget_int.BoundedIntText,
'Jupyter.Box': ipywidgets.widgets.widget_box.Box,
'Jupyter.Button': ipywidgets.widgets.widget_button.Button,
'Jupyter.Checkbox': ipywidgets.widgets.widget_bool.Checkbox,
'Jupyter.ColorPicker': ipywidgets.widgets.widget_color.ColorPicker,
'Jupyter.Controller': ipywidgets.widgets.widget_controller.Controller,
'Jupyter.ControllerAxis': ipywidgets.widgets.widget_controller.Axis,
'Jupyter.ControllerButton': ipywidgets.widgets.widget_controller.Button,
'Jupyter.Dropdown': ipywidgets.widgets.widget_selection.Dropdown,
'Jupyter.FlexBox': ipywidgets.widgets.widget_box.FlexBox,
'Jupyter.FloatProgress': ipywidgets.widgets.widget_float.FloatProgress,
'Jupyter.FloatRangeSlider': ipywidgets.widgets.widget_float.FloatRangeSlider,
'Jupyter.FloatSlider': ipywidgets.widgets.widget_float.FloatSlider,
'Jupyter.FloatText': ipywidgets.widgets.widget_float.FloatText,
'Jupyter.HTML': ipywidgets.widgets.widget_string.HTML,
'Jupyter.Image': ipywidgets.widgets.widget_image.Image,
'Jupyter.IntProgress': ipywidgets.widgets.widget_int.IntProgress,
'Jupyter.IntRangeSlider': ipywidgets.widgets.widget_int.IntRangeSlider,
'Jupyter.IntSlider': ipywidgets.widgets.widget_int.IntSlider,
'Jupyter.IntText': ipywidgets.widgets.widget_int.IntText,
'Jupyter.Label': ipywidgets.widgets.widget_string.Label,
'Jupyter.PlaceProxy': ipywidgets.widgets.widget_box.PlaceProxy,
'Jupyter.Play': ipywidgets.widgets.widget_int.Play,
'Jupyter.Proxy': ipywidgets.widgets.widget_box.Proxy,
'Jupyter.RadioButtons': ipywidgets.widgets.widget_selection.RadioButtons,
'Jupyter.Select': ipywidgets.widgets.widget_selection.Select,
'Jupyter.SelectMultiple': ipywidgets.widgets.widget_selection.SelectMultiple,
'Jupyter.SelectionSlider': ipywidgets.widgets.widget_selection.SelectionSlider,
'Jupyter.Tab': ipywidgets.widgets.widget_selectioncontainer.Tab,
'Jupyter.Text': ipywidgets.widgets.widget_string.Text,
'Jupyter.Textarea': ipywidgets.widgets.widget_string.Textarea,
'Jupyter.ToggleButton': ipywidgets.widgets.widget_bool.ToggleButton,
'Jupyter.ToggleButtons': ipywidgets.widgets.widget_selection.ToggleButtons,
'Jupyter.Valid': ipywidgets.widgets.widget_bool.Valid,
'jupyter.DirectionalLink': ipywidgets.widgets.widget_link.DirectionalLink,
'jupyter.Link': ipywidgets.widgets.widget_link.Link}
I was in a funk!
Not long after joining the University of Sheffield, I had helped convince a raft of lecturers to switch to using the Jupyter notebook for their lecturing. It was an easy piece of salesmanship and a whole lot of fun to do. Lots of people were excited by the possibilities.
The problem was that the University managed desktop was incapable of supporting an instance of the notebook with all of the bells and whistles included. As a cohort, we needed support for Python 2 and 3 kernels as well as R and even Julia. The R install needed dozens of packages and support for bioconductor. We needed LateX support to allow export to pdf and so on. We also needed to keep up to date because Jupyter development moves pretty fast! When all of this was fed into the managed desktop packaging machinery, it died. They could give us a limited, basic install but not one with batteries included.
I wanted those batteries!
In the early days, I resorted to strange stuff to get through the classes but it wasn’t sustainable. I needed a miracle to help me deliver some of the promises I had made.
Miracle delivered – SageMathCloud
During the kick-off meeting of the OpenDreamKit project, someone introduced SageMathCloud to the group. This thing had everything I needed and then some! During that presentation, I could see that SageMathCloud would solve all of our deployment woes as well as providing some very cool stuff that simply wasn’t available elsewhere. One killer-application, for example, was Google-docs-like collaborative editing of Jupyter notebooks.
I fired off a couple of emails to the lecturers I was supporting (“Everything’s going to be fine! Trust me!”) and started to learn how to use the system to support a course. I fired off dozens of emails to SageMathCloud’s excellent support team and started working with Dr Marta Milo on getting her Bioinformatics course material ready to go.
TL; DR: The course was a great success and a huge part of that success was the SageMathCloud platform
Giving back – A tutorial for lecturers on using SageMathCloud
I’m currently working on a tutorial for lecturers and teachers on how to use SageMathCloud to support a course. The material is licensed CC-BY and is available at https://github.com/mikecroucher/SMC_tutorial
If you find it useful, please let me know. Comments and Pull Requests are welcome.
I recently found myself in need of a portable install of the Jupyter notebook which made use of a portable install of R as the compute kernel. When you work in institutions that have locked-down managed Windows desktops, such portable installs can be a life-saver! This is particularly true when you are working with rapidly developing projects such as Jupyter and IRKernel.
It’s not perfect but it works for the fairly modest requirements I had for it. Here are the steps I took to get it working.
Download and install Portable Python
I downloaded Portable Python 2.7.6.1 from http://portablepython.com/ and installed into a directory called Portable Python 2.7.6.1
Update IPython and install the extra modules we need
This version of Portable Python comes with a portable IPython instance but it is too old to support alternative kernels. As such, we need to install a newer version.
Open a cmd.exe command prompt and navigate to Portable Python 2.7.6.1\App\Scripts.
Enter the command
easy_install ipython.exe
You’ll now find that you can launch the ipython.exe terminal from within this directory:
C:\Users\walkingrandomly\Desktop\Portable Python 2.7.6.1\App\Scripts>ipython Python 2.7.6 (default, Nov 10 2013, 19:24:18) [MSC v.1500 32 bit (Intel)] Type "copyright", "credits" or "license" for more information. IPython 3.1.0 -- An enhanced Interactive Python. ? -> Introduction and overview of IPython's features. %quickref -> Quick reference. help -> Python's own help system. object? -> Details about 'object', use 'object??' for extra details. In [1]: exit()
If you try to launch the notebook, however, you’ll get error messages. This is because we haven’t taken care of all the dependencies. Let’s do that now. Ensuring you are still in the Portable Python 2.7.6.1\App\Scripts folder, execute the following commands.
easy_install pyzmq easy_install jinja2 easy_install tornado easy_install jsonschema
You should now be able to launch the notebook using
ipython notebook
Install portable R and IRKernel
- I downloaded Portable R 3.2 from http://sourceforge.net/projects/rportable/files/ and installed into a directory called R-Portable
- Move this directory into the Portable Python directory. It needs to go inside Portable Python 2.7.6.1\App (see this discussion to learn how I discovered that this location was the correct one)
- Launch the Portable R executable which should be at Portable Python 2.7.6.1\App\R-Portable\R-portable.exe and install the IRKernel packages by doing
install.packages(c("rzmq","repr","IRkernel","IRdisplay"), repos="http://irkernel.github.io/")
Install additional R packages
The version of Portable R I used didn’t include various necessary packages. Here’s how I fixed that.
- Launch the Portable R executable which should be at Portable Python 2.7.6.1\App\R-Portable\R-portable.exe and install the following packages
install.packages('digest') install.packages('uuid') install.packages('base64enc') install.packages('evaluate') install.packages('jsonlite')
Install the R kernel file
Create the directory structure Portable Python 2.7.6.1\App\share\jupyter\kernels\R_kernel
Create a file called kernel.json that contains the following
{"argv": ["R-Portable/App/R-Portable/bin/i386/R.exe","-e","IRkernel::main()",
"--args","{connection_file}"],
"display_name":"Portable R"
}
This file needs to go in the R_kernel directory created earlier. Note that the kernel location specified in kernel.json uses Linux style forward slashes in the path rather than the backslashes that Windows users are used to. I found that this was necessary for the kernel to work –it was ignored by the notebook otherwise.
Finishing off
Everything created so far, including R, is in the folder Portable Python 2.7.6
I created a folder called PortableJupyter and put the Portable Python 2.7.6 folder inside it. I also created the folder PortableJupyter\notebooks to allow me to carry my notebooks around with the software that runs them.
There is a bug in Portable Python 2.7.6.1 relating to scripts like IPython.exe that have been installed using easy_install. In short, they stop working if you move the directory they’re installed in – breaking portability somewhat! (Details here)
The workaround is to launch Ipython by running the script Portable Python 2.7.6.1\App\Scripts\ipython-script.py
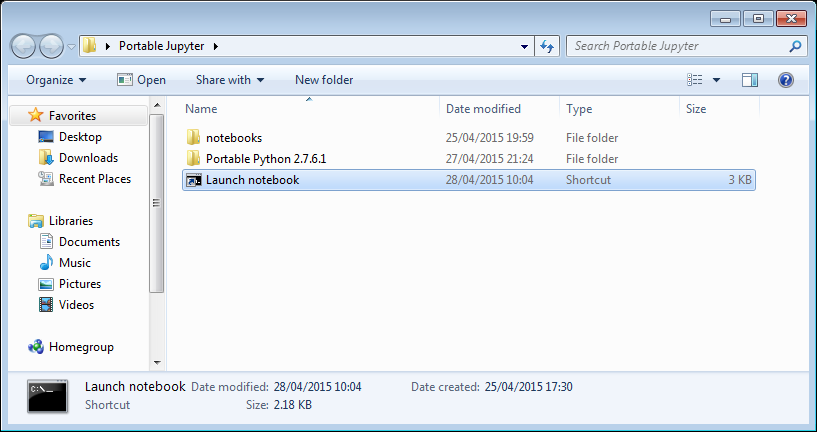
I didn’t want to bother with that so created a shortcut in my PortableJupyter folder called Launch notebook. The target of this shortcut was the following line
%windir%\system32\cmd.exe /c "cd notebooks && "%CD%/Portable Python 2.7.6.1/App\python.exe" "%CD%/Portable Python 2.7.6.1\App\Scripts\ipython-script.py" notebook"
This starts the notebook using the default web browser and puts you in the notebooks directory.
The pay off
My folder looks like this:
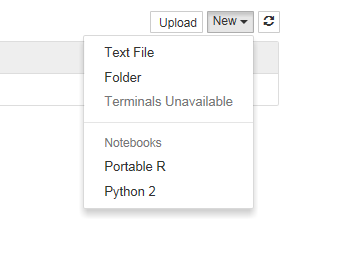
If I click on the Launch Notebook shortcut, I get a Jupyter session with 2 kernel options
I can choose the Portable R kernel and start using R in the notebook!
Many engineering textbooks such as Ogata’s Modern Control Engineering include small code examples written in languages such as MATLAB. If you don’t have access to MATLAB and if the examples don’t run in GNU Octave for some reason, the value of these textbooks is reduced.
Professor Kannan M. Moudgalya et al of the Indian Institute of Technology Bombay have developed an ambitious project that has ported the code examples of over 400 textbooks to the open-source computational system, Scilab.
The Textbook Companion Project has free Scilab code for textbooks from a range of subject areas including Fluid Mechanics, Control Systems, Chemical Engineering and Digital Electronics.
I currently work at The University of Manchester in the UK as a ‘Scientific Applications Support Specialist’. In recent years, I have noticed a steady increase in the use of open source software for both teaching and research – something that I regard as a Good Thing.
Even though Manchester has, what I believe is, a world-class site licensed software portfolio, researchers, lecturers and students often prefer open source solutions for all sorts of reasons. For example, researchers at Manchester can use MATLAB while they are associated with the University but their right to do so ceases as soon as they leave. If all of your research code is in the form of MATLAB and Simulink models, you had better hope that your next employer or school has the requisite licenses.
This summer, a few people in the Control Systems Centre of Manchester’s Electrical and Electronic Engineering department asked the question ‘Is it possible to implement all of the simple MATLAB/Simulink examples we use in a second year undergraduate introduction to Control Theory using free software?’ In particular, they chose the programs Scilab and Xcos.
Since the aim of this course is to teach control theory principles rather than any particular software solution, it would ideally be software agnostic. Students aren’t asked to develop models, they are just asked to play with pre-packaged models in order to improve their understanding of the material.
Student intern Danail Stoychev was tasked with attempting to port all of the examples from the course and in fairly short order he determined that the answer to their question was a resounding ‘Yes’.
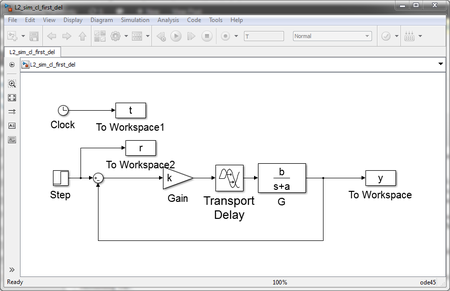
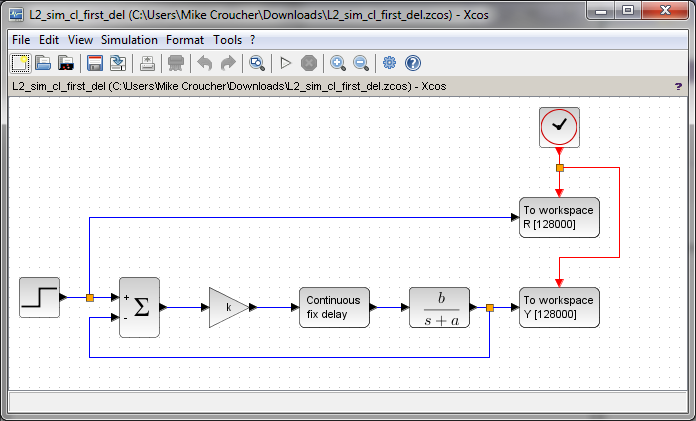
For example, the model below is an example of feedback with a first order transfer function and a delay. First in Simulink:
and now in xcos
Part of the exercise set for the students is to define all of the relevant parameters in the workspace: b,a,k and so on. If you attempt to download and run the above, you’ll have to do that in order to make them work. You’ll also need extract and plot the results from the workspace.
It can be seen that the two models look very similar and, for these examples at least, it really doesn’t matter which piece of software the students use.
The full set of MATLAB/Simulink examples along with Danail’s Scilab/Xcos conversions can be found at http://personalpages.manchester.ac.uk/staff/William.Heath/matlab_scilab.html
Earlier this year I was awarded a fellowship from the software sustainability institute, an organization that works to improve all aspects of research software. During their recent collaborations workshop in Oxford, it occurred to me that I was aware of only a relatively tiny number of software projects at my own institution, The University of Manchester. I decided to change that and started contacting our researchers to see what software they had released freely to the world as part of their research activities.
Research software comes in many forms; from small but useful MATLAB, Python or R scripts with just a handful of users and one developer right through to fully-fledged applications used by large communities of researchers and supported by teams of specialist developers. I’m interested in knowing about all of it. After all, we live in a time when even a mistake in an Excel spreadsheet can change the world.
The list below is what’s been sent to me so far and is a mirror of an internal list that’s been doing the rounds at Manchester. I’ll update it as more information becomes available. If you are at Manchester and know of a project that I’ve missed, feel free to contact me.
Last updated: 26th Jan 2015
Centre for Imaging Sciences
- BoneFinder – BoneFinder is a fully automatic segmentation tool to extract bone contours from 2D radiographs. It is written in C++ and is available for Linux and Windows.
Faculty of Life Sciences
- antiSMASH – Genome annotation tool for secondary metabolite gene clusters.
- MultiMetEval – Flux-balance analysis tool for comparative and multi-objective analysis of genome-scale metabolic models.
- mzMatch/mzmatch.R/mzMatch.ISO – Comprehensive LC/MS metabolomics data processing toolbox.
- Rank Products – Statistical tool for the identification of differentially expressed entities in molecular profiles.
Health Informatics
- openCDMS – The openCDMS project is a community effort to develop a robust, commercial-grade, full-featured and open source clinical data management system for studies and trials.
IT Services
- idiffh – Research software can produce huge text files (e.g. logs). The GNU diff program needs to read the files into memory and therefore has an upper bound on file size. idiffh might only use a simple heuristic but is only bounded by the maximum file size (and free file store).
- nearest_correlation – Python versions of nearest correlation matrix algorithms
- ParaFEM – A portable library for parallel finite element analysis. Contributions from MACE, SEAES, School of Materials.
- Shadow – This is an Apple Mac OS X shell level application that can monitor Dropbox shared folders for file deletions and restore them.
- The Reality Grid Steering Library – A software library for steering and monitoring numerical simulations, APIs available for Fortran/C++/Java and steering clients available for installation on laptops and mobile devices. Developed in collaboration with the School of computer science.
Manchester Institute of Biotechnology
- Copasi – COPASI is a software application for simulation and analysis of biochemical networks and their dynamics.
- Condor Copasi – Condor-COPASI is a web-based interface for integrating COPASI with the Condor High Throughput Computing (HTC) environment.
School of Chemical Engineering & Analytical Science
- SurfaceSpectra Identity– is free software that allows you to view and export isotope patterns.
School of Chemistry
- DOSY Toolbox – A free, open source programme for processing PFG NMR diffusion data (a.k.a. DOSY data).
- Clinical NERC – Clinical NERC is a simple customizable state-of-the-art named entity recognition, and classification software for clinical concepts or entities.
- EasyChair – EasyChair is a free conference management system.
- GPC – The University of Manchester GPC library is a flexible and highly robust polygon set operations library for use with C, C#, Delphi, Java, Perl, Python, Haskell, Lua, VB.Net (and other) applications.
- HiPLAR – High Performance Linear Algebra in R. A collaboration between Manchester and Imperial.
- iProver – 7 times word champion in theorem proving.
- INSEE – Interconnection Networks Simulation and Evaluation Environment
- KUPKB (The Kidney & Urinary Pathway Knowledge Base) – The KUPKB is a collection of omics datasets that have been extracted from scientific publications and other related renal databases. The iKUP browser provides a single point of entry for you to query and browse these datasets.
- ManTIME – ManTIME is an open-source machine learning pipeline for the extraction of temporal expressions from general domain texts.
- MethodBox – MethodBox provides a simple, easy to use environment for browsing and sharing surveys, methods and data.
- myExperiment – myExperiment makes it easy to find, use and share scientific workflows and other Research Objects, and to build communities.
- Open PHACTS Discovery Platform – Freely available, this platform integrates pharmacological data from a variety of information resources and provides tools and services to question this integrated data to support pharmacological research.
- OWL API – A Java API and reference implementation for creating, manipulating and serialising OWL Ontologies. The latest version of the API is focused towards OWL 2. The OWL API is open source and is available under either the LGPL or Apache Licenses.
- OWL Tools – a collection of tools for working with OWL ontologies
- OWL Webapps – a collection of web apps for working with OWL ontologies
- RightField – Semantic annotation by stealth. RightField is tool for adding ontology term selection to Excel spreadsheets to create templates which are then reused by Scientists to collect and annotate their data without any need to understand, or even be aware of, RightField or the ontologies used. Later the annotations can be collected as RDF
- SEEK – SEEK is a web-based platform, with associated tools, for finding, sharing and exchanging Data, Models and Processes in Systems Biology.
- ServiceCatalographer – ServiceCatalographer is an open-source Web-based platform for describing, annotating, searching and monitoring REST and SOAP Web services.
- Simple Spreadsheet Extractor – A simple ruby gem that provides a facility to read an XLS or XLSX Excel spreadsheet document and produce an XML representation of its content.
- Taverna – Taverna is an open source and domain-independent Workflow Management System – a suite of tools used to design and execute scientific workflows and aid in silico experimentation.
- TERN – TERN is a temporal expressions identification and normalisation software; designed for clinical data.
- Utopia Documents – Utopia Documents brings a fresh new perspective to reading the scientific literature, combining the convenience and reliability of the PDF with the flexibility and power of the web.
School of Earth, Atmospheric and Environmental Sciences
- ManUniCast – iPad/iPhone app. Weather and Air-Quality Forecasts for the UK and Europe
School of Electrical and Electronic Engineering
- Automatic classification of eye fixations – Identify fixations and saccades from point-of-gaze data without parametric assumptions or expert judgement. MATLAB code.
- Bootstrap Threshold Software – Estimate a threshold and a robust SD from stimulus-response data with a normal cumulative distribution. Written in C.
- LDLTS – Laplace transform Transient Processor and Deep Level Spectroscopy. A collaboration between Manchester and the Institute of Physics Polish Academy of Sciences in Warsaw
- Model-Free Psychometric Function Software – Fit a stimulus-response curve and estimate a threshold and SD without a parametric model
- Raspbian – Raspbian is a free operating system based on Debian optimized for the Raspberry Pi hardware.
- Signal Wizard – Digital signal processing software.
School of Mathematics
- EIDORS – Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software.
- Fractional Matrix Powers – MATLAB functions to compute fractional matrix powers with Frechet derivatives and condition number estimate
- fAbcond – Python code for the condition number of a matrix exponential times a vector
- funm_quad – Quadrature-based Arnoldi restarts for matrix function computations in MATLAB
- IFISS – IFISS is a graphical package for the interactive numerical study of incompressible flow problems which can be run under Matlab or Octave.
- MARKOVFUNMV – An adaptive black-box rational Arnoldi method for the approximation of Markov functions.
- Matrix Computation Toolbox – The Matrix Computation Toolbox is a collection of MATLAB M-files containing functions for constructing test matrices, computing matrix factorizations, visualizing matrices, and carrying out direct search optimization.
- Matrix Function Toolbox – The Matrix Function Toolbox is a MATLAB toolbox connected with functions of matrices.
- Matrix Logarithm – MATLAB Files. Two functions for computing the matrix logarithm by the inverse scaling and squaring method.
- Matrix Logarithm with Frechet Derivatives and Condition Number – MATLAB files
- NLEVP A Collection of Nonlinear Eigenvalue Problems – This MATLAB Toolbox provides a collection of nonlinear eigenvalue problems.
- oomph-lib – An object-oriented, open-source finite-element library for the simulation of multiphysics problems.
- rktoolbox – A Rational Krylov Toolbox for MATLAB
- Shrinking (MATLAB) – MATLAB codes for restoring definiteness of a symmetric matrix by shrinking
- Shrinking (Python) – Python codes for restoring definiteness of a symmetric matrix by shrinking
- Simfit – Free software for simulation, curve fitting, statistics, and plotting.
- SmallOverlap – SmallOverlap is a GAP 4 package which implements new, highly efficient algorithms for computing with finitely presented semigroups and monoids whose defining presentations satisfy small overlap conditions (in the sense of J.H.Remmers)
- Symmetric eigenvalue decomposition and the SVD – MATLAB files
- testing_matrix_functions – MATLAB files for testing matrix function algorithms using identities such as exp(log(A)) = A
School of Mechanical, Aerospace and Civil Engineering (MACE)
- DualSPHysics – DualSPHysics is based on the Smoothed Particle Hydrodynamics model named SPHysics and makes use of GPUs.
- FLIGHT – FLIGHT specialises in the prediction and modelling of fixed-wing aircraft performance
- SPHYSICS – SPHysics is a platform of Smoothed Particle Hydrodynamics (SPH) codes inspired by the formulation of Monaghan (1992) developed jointly by researchers at the Johns Hopkins University (U.S.A.), the University of Vigo (Spain), the University of Manchester (U.K.) and the University of Rome La Sapienza (Italy).
- SWAB Online – Innovative and User Friendly Web Application in Running Fortran-based 1-D Shallow Water near Shore Wave Simulation Modelling
School of Physics and Astronomy
- Herwig++ – Herwig++ is a new event generator, written in C++, built on the experience gained with the well-known event generator HERWIG, which was used by the particle physics community for nearly 30 years. Herwig++ is used by the LHC experiments to predict the results of their collisions and as an essential component of their data analysis. It is developed by a consortium of four main nodes, including Manchester, and its published write-up has been cited over 500 times.
- im3shape – Im3shape measures the shapes of galaxies in astronomical survey images, taking into account that they have been distorted by a point-spread function.
- MAD8/madinput – Mathematica code and MAD8 installer for performing optics calculations for particle accelerator design.
- PolyParticleTracker – MATLAB code for particle tracking against complex optical backgrounds
Simulink from The Mathworks is widely used in various disciplines. I was recently asked to come up with a list of alternative products, both free and commercial.
Here are some alternatives that I know of:
- MapleSim – A commercial Simuink replacement from the makers of the computer algebra system, Maple
- OpenModelica -An open-source Modelica-based modeling and simulation environment intended for industrial and academic usage
- Wolfram SystemModeler – Very new commercial product from the makers of Mathematica. Click here for Wolfram’s take on why their product is the best.
- xcos – This free Simulink alternative comes with Scilab.
I plan to keep this list updated and, eventually, include more details. Comments, suggestions and links to comparison articles are very welcome. If you have taught a course using one of these alternatives and have experiences to share, please let me know. Similarly for anyone who was switched (or attempted to switch) their research from Simulink. Either comment to this post or contact me directly.
I’ve nothing against Simulink but would like to get a handle on what else is out there.
There are many ways to benchmark an Android device but the one I have always been most interested in is the Linpack for android benchmark by GreeneComputing. The Linpack benchmarks have been used for many years by supercomputer builders to compare computational muscle and they form the basis of the Top 500 list of supercomputers.
Linpack measures how quickly a machine can solve a dense n by n system of linear equations which is a common task in scientific and engineering applications. The results of the benchmark are measured in flops which stands for floating point operations per second. A typical desktop PC might acheive around 50 gigaflops (50 billion flops) whereas the most powerful PCs on Earth are measured in terms of petaflops (Quadrillions of flops) with the current champion weighing in at 16 petaflops, that’s 16,000,000,000,000,000 floating point operations per second–which is a lot!
Acording to the Android Linpack benchmark, my Samsung Galaxy S2 is capable of 85 megaflops which is pretty powerful compared to supercomputers of bygone eras but rather weedy by today’s standards. It turns out, however, that the Linpack for Android app is under-reporting what your phone is really capable of. As the authors say ‘This test is more a reflection of the state of the Android Dalvik Virtual Machine than of the floating point performance of the underlying processor.’ It’s a nice way of comparing the speed of two phones, or different firmwares on the same phone, but does not measure the true performance potential of your device.Put another way, it’s like measuring how hard you can punch while wearing huge, soft boxing gloves.
Rahul Garg, a PhD. student at McGill University, thought that it was high time to take the gloves off!
rgbench – a true high performance benchmark for android devices
Rahul has written a new benchmark app called RgbenchMM that aims to more accurately reflect the power of modern Android devices. It performs a different calculation to Linpack in that it meaures the speed of matrix-matrix multiplication, another common operation in sicentific computing.
The benchmark was written using the NDK (Native Development Kit) which means that it runs directly on the device rather than on the Java Virtual Machine, thus avoiding Java overheads. Furthermore, Rahul has used HPC tricks such as tiling and loop unrolling to squeeze out the very last drop of performance from your phone’s processor . The code tests about 50 different variations and the performance of the best version found for your device is then displayed.
When I ran the app on my Samsung Galaxy S2 I noted that it takes rather longer than Linpack for Android to execute – several minutes in fact – which is probably due to the large number of variations its trying out to see which is the best. I received the following results
- 1 thread: 389 Mflops
- 2 threads: 960 Mflops
- 4 threads: 867.0 Mflops
Since my phone has a dual core processor, I expected performance to be best for 2 threads and that’s exactly what I got. Almost a Gigaflop on a mobile phone is not bad going at all! For comparison, I get around 85 Mflops on Linpack for Android. Give it a try and see how your device compares.

Links
- RgbenchMM on GooglePlay
- Prelim Analysis of RgbenchMM – Some of the in-depth details of the benchmark, written by the app’s author.
- Supercomputers vs mobile phones