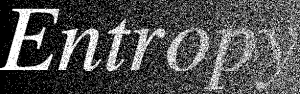Archive for the ‘University of Sheffield’ Category
My friends over at the University of Sheffield Research Software Engineering group are running a GPU Hackathon sponsored by Nvidia. The event will be on August 19-23 2019 in Sheffield, United Kingdom. The call for proposals is at http://gpuhack.shef.ac.uk/
The Sheffield team have this to say about the event:
We are looking for teams of 3-5 developers with a scalable** application to port to or optimize on a GPU accelerator. Collectively the team must have complete knowledge of the application. If the application is a suite of apps, no more than two per team will be allowed and a minimum of 2 people per app must attend. Space will be limited to 8 teams.
** By scalable we mean node-to-node communication implemented, but don’t be discouraged from applying if your application is less than scalable. We are also looking for breadth of application areas.
The goal of the GPU hackathon is for current or prospective user groups of large hybrid CPU-GPU systems to send teams of at least 3 developers along with either:
- (potentially) scalable application that could benefit from GPU accelerators, or
- An application running on accelerators that needs optimization.
There will be intensive mentoring during this 5-day hands-on workshop, with the goal that the teams leave with applications running on GPUs, or at least with a clear roadmap of how to get there.
I work at The University of Sheffield where I am one of the leaders of the new Research Software Engineering function. One of the things that my group does is help people make use of Sheffield’s High Performance Computing cluster, Iceberg.
Iceberg is a heterogenous system with around 3440 CPU cores and a sprinkling of GPUs. It’s been in use for several years and has been upgraded a few times over that period. It’s a very traditional HPC system that makes use of Linux and a variant of Sun Grid Engine as the scheduler and had served us well.
A while ago, the sysadmin pointed me to a goldmine of a resource — Iceberg’s accounting log. This 15 Gigabyte file contains information on every job submitted since July 2009. That’s more than 7 years of the HPC usage of 3249 users — over 46 million individual jobs.
The file format is very straightforward. There’s one line per job and each line consists of a set of colon separated fields. The first few fields look like something like this:
long.q:node54.iceberg.shef.ac.uk:el:abc07de:
The username is field 4 and the number of slots used by the job is field 35. On our system, slots correspond to CPU cores. If you want to run a 16 core job, you ask for 16 slots.
With one line of awk, we can determine the maximum number of slots ever requested by each user.
gawk -F: '$35>=slots[$4] {slots[$4]=$35};END{for(n in slots){print n, slots[n]}}' accounting > ./users_max_slots.csv
As a quick check, I grepped the output file for my username and saw that the maximum number of cores I’d ever requested was 20. I ran a 32 core MPI ‘Hello World’ job, reran the line of awk and confirmed that my new maximum was 32 cores.
There are several ways I could have filtered the number of users but I was having awk lessons from David Jones so let’s create a new file containing the users who have only ever requested 1 slot.
gawk -F: '$35>=slots[$4] {slots[$4]=$35};END{for(n in slots){if(slots[n]==1){print n, slots[n]}}}' accounting > users_where_max_is_one_slot.csv
Running wc on these files allows us to determine how many users are in each group
wc users_max_slots.csv 3250 6498 32706 users_max_slots.csv
One of those users turned out to be a blank line so 3249 usernames have been used on Iceberg over the last 7 years.
wc users_where_max_is_one_slot.csv 2393 4786 23837 users_where_max_is_one_slot.csv
That is, 2393 of our 3249 users (just over 73%) over the last 7 years have only ever run 1 slot, and therefore 1 core, jobs.
High Performance?
So 73% of all users have only ever submitted single core jobs. This does not necessarily mean that they have not been making use of parallelism. For example, they might have been running job arrays – hundreds or thousands of single core jobs performing parameter sweeps or monte carlo simulations.
Maybe they were running parallel codes but only asked the scheduler for one core. In the early days this would have led to oversubscribed nodes, possibly up to 16 jobs, each trying to run 16 cores.These days, our sysadmin does some voodoo to ensure that jobs can only use the number of cores that have been requested, no matter how many threads their code is spawning. Either way, making this mistake is not great for performance.
Whatever is going on, this figure of 73% is surprising to me!
Thanks to David Jones for the awk lessons although if I’ve made a mistake, it’s all my fault!
Update (11th Jan 2017)
UCL’s Ian Kirker took a look at the usage of their general purpose cluster and found that 71.8% of their users have only ever run 1 core jobs. https://twitter.com/ikirker/status/819133966292807680
I was recently invited to give a talk at the Sheffield R Users Group and decided to give a brief overview of how R relates to other technologies. Subjects included Mathematica’s integration of R, Intel’s compilers, Math Kernel Library and how they can make R faster and a range of Microsoft technologies including R Tools for Visual Studio, Microsoft R Open and the MRAN for reproducibility. I also touched upon the NAG Library, Maple’s code generation for R, GPUs and Spark.
- Slide deck: _ and R: How R relates to other technologies
Did I miss anything? If you were to give a similar talk, what might you have included?
One of the great things about being a Research Software Engineer is the diversity of work you can get involved with. I specialise in smaller interventions which means that I can be working with physicists on Monday, engineers on Tuesday, geneticists on Wednesday….you get the idea.
Last month, I got to work with some Ecologists along with Anna Krystalli. We undertook the arduous journey from Sheffield down to Exeter to deliver talks and workshops at a post-conference symposium on reproducibility in science, organised by Malika Ihle and Isabel Winney, at the International Symposium on Behavioural Ecology.
I gave my talk, Is your research software correct?, and also delivered a workshop on using projects and version control using R and RStudio in the Code Cafe style. For the full write up of the day, see the excellent blog post by Anna over at the Mozilla Science Lab blog.
Updates : More resources
- Elevating the status of code in Ecology. Thanks to @katerererena for pointing this one out to me.
- Anna Krystalli’s course material
I was in a funk!
Not long after joining the University of Sheffield, I had helped convince a raft of lecturers to switch to using the Jupyter notebook for their lecturing. It was an easy piece of salesmanship and a whole lot of fun to do. Lots of people were excited by the possibilities.
The problem was that the University managed desktop was incapable of supporting an instance of the notebook with all of the bells and whistles included. As a cohort, we needed support for Python 2 and 3 kernels as well as R and even Julia. The R install needed dozens of packages and support for bioconductor. We needed LateX support to allow export to pdf and so on. We also needed to keep up to date because Jupyter development moves pretty fast! When all of this was fed into the managed desktop packaging machinery, it died. They could give us a limited, basic install but not one with batteries included.
I wanted those batteries!
In the early days, I resorted to strange stuff to get through the classes but it wasn’t sustainable. I needed a miracle to help me deliver some of the promises I had made.
Miracle delivered – SageMathCloud
During the kick-off meeting of the OpenDreamKit project, someone introduced SageMathCloud to the group. This thing had everything I needed and then some! During that presentation, I could see that SageMathCloud would solve all of our deployment woes as well as providing some very cool stuff that simply wasn’t available elsewhere. One killer-application, for example, was Google-docs-like collaborative editing of Jupyter notebooks.
I fired off a couple of emails to the lecturers I was supporting (“Everything’s going to be fine! Trust me!”) and started to learn how to use the system to support a course. I fired off dozens of emails to SageMathCloud’s excellent support team and started working with Dr Marta Milo on getting her Bioinformatics course material ready to go.
TL; DR: The course was a great success and a huge part of that success was the SageMathCloud platform
Giving back – A tutorial for lecturers on using SageMathCloud
I’m currently working on a tutorial for lecturers and teachers on how to use SageMathCloud to support a course. The material is licensed CC-BY and is available at https://github.com/mikecroucher/SMC_tutorial
If you find it useful, please let me know. Comments and Pull Requests are welcome.
I learned about entropy as part of my undergraduate Physics education but it turns out that the concept of entropy turns up in many fields including linguistics, themodynamics, information theory, chemistry and artificial intelligence.
As part of Sheffield’s Open Data Science Initiative, computer scientist, Neil Lawrence, has teamed up with linguist, Dagmar Divjak, to organise a cross-faculty discussion meeting on the subject of entropy.
For more details on the day’s events, and to register, see http://opendsi.cc/ed2016/program
I wasted a little time producing the above logo for the event using Mathematica.
Here’s the source code:-
(*consider column one pixel at a time. Invert the pixel if a random number is below some threshold*)
flipbit[col_, prob_] := Module[{result, temp, x},
result = col;
Do[
If[RandomReal[] <= prob,
If[result[[x]] == 1, result[[x]] = 0, result[[x]] = 1];
]
, {x, 1, Length[col]}
];
Return[result]
]
text = "Entropy";
image = Rasterize[Text[Style[text, White, Italic, 190]],
Background -> Black];
imageData = ImageData[Binarize[image]];
const = 1/Dimensions[imageData][[2]]*0.42;
(*Apply flipbit to all columns. Increase probability of flipping as you move along the x-axis*)
logo =
Transpose[
MapIndexed[flipbit[#1, const*#2[[1]]] &, Transpose[imageData]]];
Image[logo]
Finally, I found this quote about entropy that I quite like:
You should call it entropy, for two reasons. In the first place your uncertainty function has been used in statistical mechanics under that name, so it already has a name. In the second place, and more important, no one really knows what entropy really is, so in a debate you will always have the advantage.
John von Neumann to Claude Shannon a name for his new uncertainty function. Source: Wikiquotes
The University of Sheffield recently purchased licenses for the Windows version of the Intel compiler suite and I’m involved in the release to campus process. As part of this, I wrote some basic documentation on how to compile Hello World using the Intel C++ Compiler within Visual Studio Community Edition 2015. Since this may be useful to people outside of University of Sheffield, I reproduce this part of our documentation here.
These notes were prepared using Intel C++ XE 2015 version 15.0.6 and Visual Studio Community Edition 2015 RTM version. Note that there are problems with using this version of Intel C++ and later versions of Visual Studio Community Edition.
Click on any of the images to see them full size.
- Launch Visual Studio and click on File->New Project
- Choose Visual C++ -> Win32 Console Application and give your project a name.
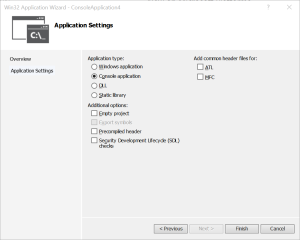
- In the Win32 Application wizard, untick Precompiled header and Security Development Lifecycle (SDL) checks and click Finish.
- A skeleton main() function will appear. Modify the code so that it reads
#include
int main()
{
std::cout << "Hello World";
return 0;
}
- Click on Project->HelloWorld Properties and under the C++ -> General section, ensure that Suppress Startup Banner is set to No and click OK. This will ensure that when you compile, you’ll be able to see that it’s the Intel Compiler doing the work rather than Visual Studio C++.
- Click on Project->Intel Compiler->Use Intel C++
- Build and run the code by pressing CTRL F5. You should see something like the following
>------ Build started: Project: HelloWorld, Configuration: Debug Win32 ------ 1> icl /Qvc14 "/Qlocation,link,C:\Program Files (x86)\Microsoft Visual Studio 14.0\VC\bin" /ZI /W3 /Od /Qftz- -D __INTEL_COMPILER=1500 -D WIN32 -D _DEBUG -D _CONSOLE -D _UNICODE -D UNICODE /EHsc /RTC1 /MDd /GS /Zc:wchar_t /Zc:forScope /FoDebug\ /FdDebug\vc140.pdb /Gd /TP HelloWorld.cpp stdafx.cpp 1> 1> Intel(R) C++ Compiler XE for applications running on IA-32, Version 15.0.6.285 Build 20151119 1> Copyright (C) 1985-2015 Intel Corporation. All rights reserved.
Once the compilation has completed, a console window should pop up showing the Hello World output.
Back in December 2014, I learned that I’d be moving from The University of Manchester to The University of Sheffield to do the type of thing I’ve always done which is a combination of research software engineering and research software support.
I’ve been in Sheffield for two months now and am having a blast! There’s so much cool stuff going on here that it makes my head spin a little and the community at Sheffield have welcomed me with open arms. It truly is a wonderful place in which to work.
One of the departments I’ve started working with is The Sheffield Institute for Translation Neuroscience (SITraN). My contributions have been relatively minor so far – A bit of Python coding for a machine learning project called GPy and some code speed-up work in R for Winston Hide and his collaborators. When I hang out in SITraN, I usually sit with the machine learning people and listen in on their conversions about Python, MATLAB, GPUs, C++ and R — it’s essentially Nerdvana for someone like me.
On to the point of this blog post. SITraN now have their own blog called SITraNsmissions where they’ll be discussing various aspects of their work and how it applies the principles of neuroscience to help treat diseases such as Motor Neurone Disease (MND). In the video below, taken from SITraNsmissions first blog post, Professor Pamela Shaw gives an overview of the work that SITraN does.