Archive for the ‘Linux’ Category
In my previous blog post I mentioned that I am a member of a team that supports High Throughput Computing (HTC) at The University of Manchester via a 1600+ core ‘condor pool’. In order to make it as easy as possible for our researchers to make use of this resource one of my colleagues, Ian Cottam, created a system called DropAndCompute. In this guest blog post, Ian describes DropAndCompute and how it evolved into the system we use at Manchester today.
The Evolution of “DropAndCompute” by Ian Cottam
DropAndCompute, as used at The University of Manchester’s Faculty of Engineering and Physical Sciences, is an approach to using network (or grid or cloud based) computational resources without having to know the operating system of the resource’s gateway or any command line tools of either the resource itself —Condor in our case — or in general. Most such gateways run a flavour of Unix, often Linux. Many of our users are either unfamiliar with Linux or just prefer a drag-and-drop interface, as I do myself despite using various flavours of Unix since Version 6 in the late 70s.
Why did I invent it? On its original web site description page wiki.myexperiment.org/index.php/DropAndCompute the following reasons are given:
- A simple and uniform drag-and-drop graphical user interface, potentially, to many resource pools.
- No use of terminal windows or command lines.
- No need to login to remote hosts or install complicated grid-enabling software locally.
- No need for the user to have an account on the remote resources (instead they are accounted by having a shared folder allocated). Of course, nothing stops the users from having accounts should that be preferred.
- No need for complicated Virtual Private Networks, IP Tunnelling, connection brokers, or similar, in order to access grid resources on private subnets (provided at least one node is on the public Internet, which is the norm).
- Pop-ups notify users of important events (basically, log and output files being created when a job has been accepted, and when the generated result files arrive).
- Somewhat increased security as the user only has (indirect) access to a small subset of the computational resource’s commands.
Version One
The first version was used on a Condor Pool within our interdisciplinary biocentre (MIB). A video of it in use is shown below
Please do take the time to look at this video as it shows clearly how, for example, Condor can be used via this type of interface.
This version was notable for using the commercial service: Dropbox and, in fact, my being a Dropbox user inspired the approach and its name. Dropbox is trivial to install on any of the main platforms, on any number of computers owned by a user, and has a free version giving 2GB of synchronised and shared storage. In theory, only the computational resource supplier need pay for a 100GB account with Dropbox, have a local Condor submitting account, and share folders out with users of the free Dropbox-based service.
David De Roure, then at the University of Southampton and now Oxford, reviewed this approach here at blog.openwetware.org/deroure/?p=97, and offers his view as to why it is important in helping scientists start on the ‘ramp’ to using what can be daunting, if powerful, computational facilities.
Version Two
Quickly the approach migrated to our full, faculty-wide Condor Pool and the first modification was made. Now we used separate accounts for each user of the service on our submitting nodes; Dropbox still made this sharing scheme trivial to set up and manage, whilst giving us much better usage accounting information. The first minor problem came when some users needed more –much more in fact– than 2GB of space. This was solved by them purchasing their own 50GB or 100GB accounts from Dropbox.
Problems and objections
However, two more serious problems impacted our Dropbox based approach. First, the large volume of network traffic across the world to Dropbox’s USA based servers and then back down to local machines here in Manchester resulted in severe bottlenecks once our Condor Pool had reached the dizzy heights of over a thousand processor cores. We could have ameliorated this by extra resources, such as multiple submit nodes, but the second problem proved to be more of a showstopper.
Since the introduction of DropAndCompute several people –at Manchester and beyond– have been concerned about research data passing through commercial, USA-based servers. In fact, the UK’s National Grid Service (NGS) who have implemented their own flavour of DropAndCompute did not use Dropbox for this very reason. The US Patriot Act means that US companies must surrender any data they hold if officially requested to do so by Federal Government agencies. Now one approach to this is to do user-level encryption of the data before it enters the user’s dropbox. I have demonstrated this approach, but it complicates the model and it is not so straightforward to use exactly the same method on all of the popular platforms (Windows, Mac, Linux).
Version Three
To tackle the above issues we implemented a ‘local version’ of DropAndCompute that is not Dropbox based. It is similar to the NGS approach, but, in my opinion, much simpler to setup. The user merely has to mount a folder on the submit node on their local computer(s), and then use the same drag-and-drop approach to get the job initiated, debugged and run (or even killed, when necessary). This solves the above issues, but could be regarded as inferior to the Dropbox based approach in five ways:
1. The convenience and transparency of ‘offline’ use. That is, Dropbox jobs can be prepared on, say, a laptop with or without net access, and when the laptop next connects the job submissions just happens. Ditto for the results coming back.
2. When online and submitting or waiting for results with the local version, the folder windows do not update to give the user an indication of progress.
3. Users must remember to use an email notification that a job has finished, or poll to check its status.
4. The initial setup is a little harder for the local version compared with using Dropbox.
5. The computation’s result files are not copied back automatically.
So far, only item 5 has been remarked on by some of our users, and it, and the others, could be improved with some programming effort.
A movie of this version is shown below; it doesn’t have any commentary, but essentially follows the same steps as the Dropbox based video. You will see the network folder’s window having to be refreshed manually –this is necessary on a Mac (but could be scripted); other platforms may be better– and results having to be dragged back from the mounted folder.
I welcome comments on any aspect of this –still evolving– approach to easing the entry ‘cost’ to using distributed computing resources.
Acknowledgments
Our Condor Pool is supported by three colleagues besides myself: Mark Whidby, Mike Croucher and Chris Paul. Mark, inter alia, maintains the current version of DropAndCompute that can operate locally or via Dropbox. Thanks also to Mike for letting me be a guest on Walking Randomly.
I thought I’d try out the new image capturing functionality in Mathematica 8 on my Linux based laptop with built in webcam but…
In[1]:= ImageCapture[] During evaluation of In[1]:= ImageCapture::notsupported: Image acquisition is not supported on Unix. >> Out[1]= ImageCapture[]
That makes me a sad panda!
Christmas isn’t all that far away so I thought that it was high time that I wrote my Christmas list for mathematical software developers and vendors. All I want for christmas is….
Mathematica
- A built in ternary plot function would be nice
- Ship workbench with the main product please
- An iPad version of Mathematica Player
MATLAB
- Merge the parallel computing toolbox with core MATLAB. Everyone uses multicore these days but only a few can feel the full benefit in MATLAB. The rest are essentially second class MATLAB citizens muddling by with a single core (most of the time)
- Make the mex interface thread safe so I can more easily write parallel mex files
Maple
- More CUDA accelerated functions please. I was initially excited by your CUDA package but then discovered that it only accelerated one function (Matrix Multiply). CUDA accelerated Random Number Generators would be nice along with fast Fourier transforms and a bit more linear algebra.
MathCAD
- Release Mathcad Prime.
- Mac and Linux versions of Mathcad. Maple,Mathematica and MATLAB have versions for all 3 platforms so why don’t you?
NAG Library
- Produce vector versions of functions like g01bk (poisson distribution function). They might not be needed in Fortran or C code but your MATLAB toolbox desperately needs them
- A Mac version of the MATLAB toolbox. I’ve got users practically begging for it :)
- A NAG version of the MATLAB gamfit command
Octave
- A just in time compiler. Yeah, I know, I don’t ask for much huh ;)
- A faster pdist function (statistics toolbox from Octave Forge). I discovered that the current one is rather slow recently
SAGE Math
- A Locator control for the interact function. I still have a bounty outstanding for the person who implements this.
- A fully featured, native windows version. I know about the VM solution and it isn’t suitable for what I want to do (which is to deploy it on around 5000 University windows machines to introduce students to one of the best open source maths packages)
SMath Studio
- An Android version please. Don’t make it free – you deserve some money for this awesome Mathcad alternative.
SpaceTime Mathematics
- The fact that you give the Windows version away for free is awesome but registration is a pain when you are dealing with mass deployment. I’d love to deploy this to my University’s Windows desktop image but the per-machine registration requirement makes it difficult. Most large developers who require registration usually come up with an alternative mechanism for enterprise-wide deployment. You ask schools with more than 5 machines to link back to you. I want tot put it on a few thousand machines and I would happily link back to you from several locations if you’ll help me with some sort of volume license. I’ll also give internal (and external if anyone is interested) seminars at Manchester on why I think Spacetime is useful for teaching mathematics. Finally, I’d encourage other UK University applications specialists to evaluate the software too.
- An Android version please.
How about you? What would you ask for Christmas from your favourite mathematical software developers?
I recently had a set of files that were named as follows
frame1.png frame2.png frame3.png frame4.png frame5.png frame6.png frame7.png frame8.png frame9.png frame10.png
and so on, right up to frame750.png. The plan was to turn these .png files into an uncompressed movie using mencoder via the following command (original source)
mencoder mf://*.png -mf w=720:h=720:fps=25:type=png -ovc raw -oac copy -o output.avi
but I ended up with a movie that jumped all over the place since the frames were in an odd order. In the following order in fact
frame0.csv frame100.csv frame101.csv frame102.csv frame103.csv frame104.csv frame105.csv frame106.csv frame107.csv frame108.csv frame109.csv frame10.csv frame110.csv
This is because globbing expansion (the *.png bit) is alphabetical in bash rather than numerical.
One way to get the frames in the order that I want is to zero-pad them. In other words I replace file1.png with file001.png and file20.png with file020.png and so on. Here’s how to do that in bash
#!/bin/bash
num=`expr match "$1" '[^0-9]*\([0-9]\+\).*'`
paddednum=`printf "%03d" $num`
echo ${1/$num/$paddednum}
Save the above to a file called zeropad.sh and then do the following command to make it executable
chmod +x ./zeropad.sh
You can then use the zeropad.sh script as follows
./zeropad.sh frame1.png
which will return the result
frame001.png
All that remains is to use this script to rename all of the .png files in the current directory such that they are zeropadded.
for i in *.png;do mv $i `./zeropad.sh $i`; done
You may want to change the number of digits used in each filename from 3 to 5 (say). To do this just change %03d in zeropad.sh to %05d
Let me know if you find this useful or have an alternative solution you’d like to share (in another language maybe?)
I was recently asked to install 32bit Gaussian 03 binaries on an Ubuntu 9.10 machine and when I tried to run a test job I got the following error message
Erroneous write during file extend. write -1 instead of 4096 Probably out of disk space. Erroneous write during file extend. write -1 instead of 4096 Probably out of disk space. Write error in NtrExt1 Write error in NtrExt1: Bad address Segmentation fault
A bit of googling suggested that the following might work
sudo echo 0 > /proc/sys/kernel/randomize_va_space
but this will result in permission denied (explanation here). The command you really want to use is
sudo bash -c "echo 0 > /proc/sys/kernel/randomize_va_space"
Once this was done, Gaussian worked as advertised. Maybe this post will help a googler sometime in the future.
I was recently playing with some parallel code that used gfortran and OpenMP to parallelise a calculation over 4 threads and was getting some seriously odd timing results. The first time I ran the code it completed in 4.8 seconds, the next run took 4.8 seconds but the third run took 84 seconds (No, I haven’t missed off the decimal point). Subsequent timings were all over the place – 12 seconds, 4.8 seconds again, 14 seconds….something weird was going on.
I wouldn’t mind but all of these calculations had exactly the same input/output and yet sometimes this parallel code took significantly longer to execute than the serial version.
On drilling down I discovered that two of the threads had 100% CPU utilisation and the other two only had 50%. On a hunch I wondered if the CPU frequency ‘on demand’ scaling thing was messing things (It has done so in the past) up so I did
sudo cpufreq-selector -c 0 -f 3000 sudo cpufreq-selector -c 1 -f 3000 sudo cpufreq-selector -c 2 -f 3000 sudo cpufreq-selector -c 3 -f 3000
to set the cpu-frequency of all 4 cores to the maxium 3Ghz. This switched off the ‘on demand’ setting that is standard in Ubuntu.
Lo! it worked! 4.8 seconds every time. When I turned the governor back on
sudo cpufreq-selector --governor ondemand
I got back the ‘sometimes it’s fast, sometimes it’s slow’ behaviour. Oddly, one week and several reboots later I can’t get back the slow behaviour no matter what I set the governor to.
Perhaps this was just a temporary glitch in my system but, as I said earlier, I have seen this sort of behaviour before so, just to be on the safe side, it might be worth switching off the automatic governor whenever you do parallel calculations in Linux.
Does anyone have any insight into this? Comments welcomed.
The student version of MATLAB is a bargain since it only costs $99 (or less than £50 for those of us in the UK) and it comes complete with several toolboxes including symbolic maths, statistics and optimisation. Most of it is identical to the full academic version which would cost well over a thousand pounds if you bought all of the toolboxes included in the student version. The only place where it is ‘crippled’ is within Simulink since your Simulink models are limited to 1000 blocks. There are one or two other differences too and I refer you to the amazon page
for full details (Amazon is also the cheapest place I could find for Student MATLAB by the way).
One problem with the student version of MATLAB though is the fact that they only supply a 32 bit version. This was potentially a big problem for a friend of mine since he has a 64bit Linux machine with 4Gb of RAM. Fortunately, the 32bit version installs OK on 64bit Linux but the result is completely unsupported by Mathworks. Fair enough….It works and he is (mostly) happy since he gets a lot of functionality for his money.
One thing that definitely didn’t work though was compiling mex files. No matter how hard we tried we simply could not get it to work which made me look bad because I am supposed to be ‘The MATLAB guy’ around here. Well, sometimes it’s not what you know but who you know that counts and I know a LOT of MATLAB users. One of them has provided a fix but doesn’t want his name plastered all over Walking Randomly. So, thanks to Mr Anonymous we got mex files working on Student MATLAB 2009a running on 64bit Linux and this is how we did it.
Get a simple mex file and try to compile it. You’ll probably get this error.
/usr/bin/ld: cannot find -lmx
The way to get around that is to run the following command in MATLAB just before you try to compile a mex file
setenv('MATLAB_ARCH', 'glnx86') (IN MATLAB)
That gets you a little further but you’ll next be hit by
/usr/bin/ld: cannot find -lstdc++
To fix this you need to find your mexopts.sh file and change the line
CLIBS="$CLIBS -lstdc++"
to
CLIBS="$CLIBS -L/home/paul/matlab/R2009a-student/sys/os/glnx86 -lstdc++"
obviously, you’ll need to change /home/paul/matlab to wherever you actually installed MATLAB.
Your next step is to do the following in a bash prompt
ln -s /home/paul/matlab/R2009a-student/sys/os/glnx86/libstdc++.so.6 \ /home/paul/matlab/R2009a-student/sys/os/glnx86/libstdc++.so
again – substituting wherever you installed MATLAB for /home/paul/matlab
Paul was running Ubuntu 9.04 and he got the following error at some point (I can’t remember where)
/usr/include/gnu/stubs.h:7:27: error: gnu/stubs-32.h: No such file or directory
which was fixed by
sudo apt-get install libc6-dev-i386
That’s pretty much it. You should now be able to compile mex files. Hope this helps someone out there.
One of the problems with the Linux MATLAB installer is that it doesn’t add any items to the menu in windows managers such as GNOME or KDE so you have to do it manually. Now, there are several ways to do this but one of the easiest is to use the GUI tools provided with Ubuntu (assuming you are using Ubuntu of course). For the record I am using Ubuntu 9.10 (Karmic Koala) with the default GNOME windows manager along with MATLAB 2009b but I imagine that these instructions will work for a few other configurations too.
First things first, you’ll want to download a nice, scalable MATLAB icon since the ones included with MATLAB itself are a bit crude to say the least. I tried to create one directly from MATLAB using the logo command and the Scalable Vector Graphics (SVG) Export of Figures package on The File Exchange but the result wasn’t very good (see below).

Fortunately someone called malte has created a much nicer .svg file (using inkscape) that looks just like the MATLAB logo and made it available via his blog.
Once you have the svg file you can start creating the shortcut as follows:
- On the GNOME Desktop click on System->Preferences->Main Menu
- Once the Main Menu program has started choose where you want your MATLAB icon to go and click on New Item. In the screenshot below I have started to put it in the Programming submenu.
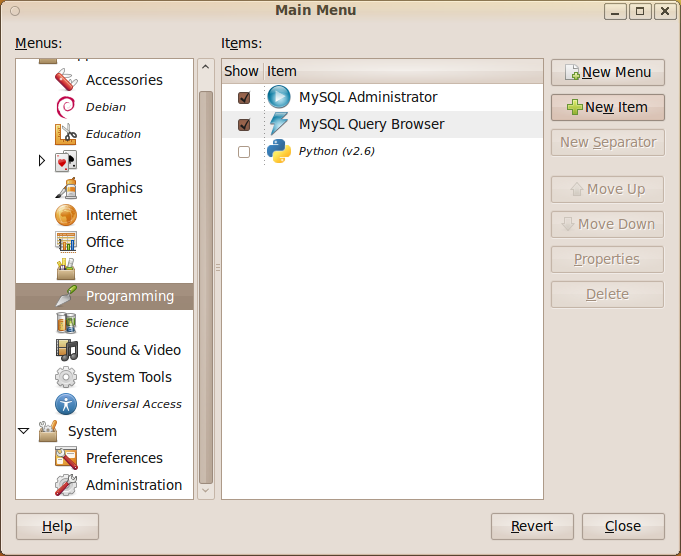
- Give your shortcut a name and put matlab -desktop in the command field. If you didn’t create a shortcut to the matlab executable when you installed the program then you may need to put the full path in instead (i.e. something like /opt/MATLAB/2009b/bin/matlab )

- Now let’s sort out the icon. Click on the spring on the left hand side of the above window to get the window below.
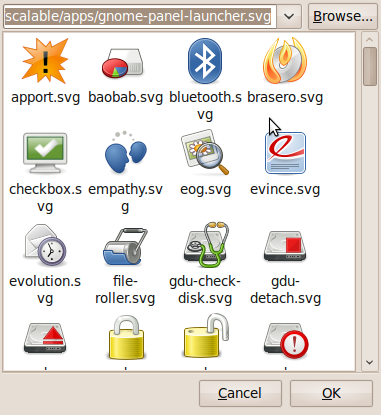
- Click on Browse and then select the folder that contains the .svg file you downloaded from malte’s blog. I put it in my Desktop folder for the screenshot below. Note that the svg file doesn’t appear in the preview pane. Click on Open.
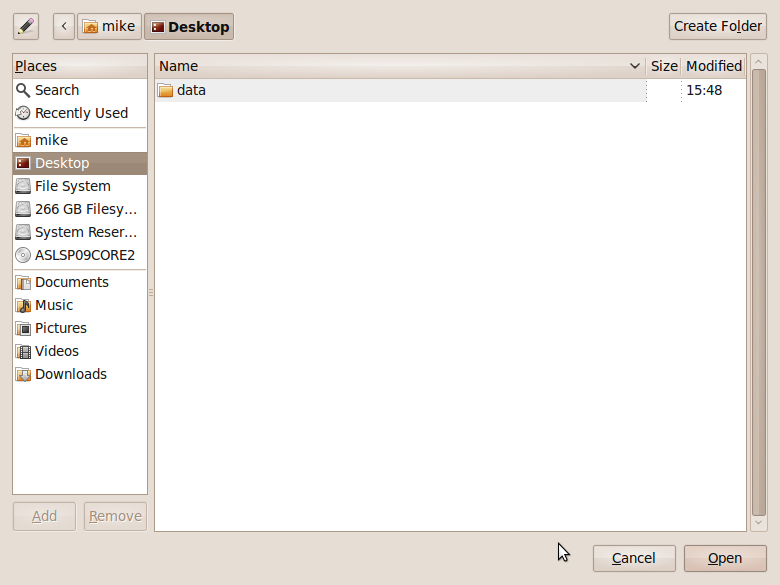
- Choose your icon.

- Click on Close and you are done
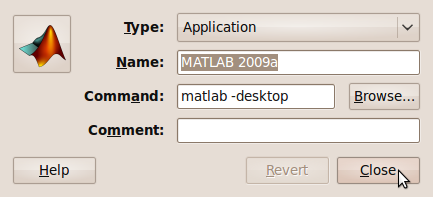
If you are using the Student version of MATLAB then the process is slightly different and has been covered by my friend Paul Brabban over at CrossedStreams.com.
I’ve just installed Ubuntu version 9.10 (Karmic Koala) from scratch on my new work machine and noticed that you can no longer right click on a file to encrypt it. The functionality is still there – it just isn’t available by default. To get it just install the seahorse-plugins package as follows
sudo apt-get install seahorse-plugins
I did a reboot to get the changes to take but there is probably a less drastic way of getting the job done.
It won’t be long before I give up Windows Mobile for good in favour of the HTC Hero and the Android operating system but before I do I thought I would take one last look at what’s out there in terms of mathematical software for the Windows Mobile platform. I last looked at this back in early 2008 but one or two things have changed since then.
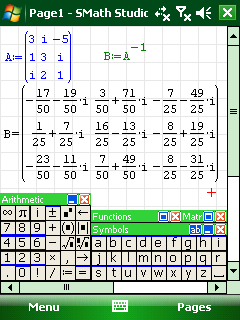
SMath studio is a very nice free maths application that is being developed by Andrey Ivashov for Windows and Linux (via Mono) desktops as well as Windows Mobile handhelds. It installed perfectly on my HTC TyTn II and offers an experience very similar to a simplified version of PTC’s Mathcad and can even handle Mathcad’s .xmcd file format in some cases. One cool feature to consider is that you can develop calculations on the desktop version (screen shot below) and then carry them around, edit and run them on your Windows Mobile phone (screenshot above).
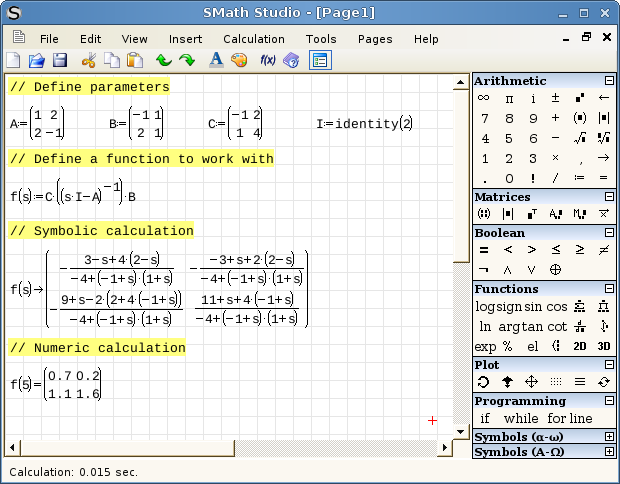
Andrey has recently released version 0.82 of the software and there are some active forums over at his website in both English and Russian where you can discuss it. This software is well worth watching in my opinion. Although I won’t be able to follow the Windows Mobile versions after next week, I’ll definitely be keeping an eye on SMath for Linux and Windows and hope to bring a more detailed review to you at some point in the future.
